Batch Loading Taxonomic Data
This page describes how taxonomy can be added to a Symbiota portal in bulk.
A user must have Super Administrator or Taxonomic Administrator permissions to add to or edit the thesaurus.
Format the Input File
Taxonomic names can be batch loaded into the thesaurus from a flat, structured comma- or tab-delimited text file. To upload an Excel file, first save as a CSV (comma delimited) text file. Formatting this file correctly is important to ensuring your taxonomy is successfully imported. At minimum, the file must include a column for ScientificName and the parent taxon (i.e. the taxon that nests immediately above the scientificName in your taxonomic hierarchy).
For example, to import the name Buccinum polaris Gray, 1839, your input file could include the following fields:
| Taxon Rank | Parent | ScientificName | Family | Author | RankID |
|---|---|---|---|---|---|
| species | Buccinum | Buccinum polaris | Buccinidae | Gray, 1839 | 220 |
However, if the higher taxonomy for this name does not already exist in your portal, the following rows should also be included in your file:
| Taxon Rank | Parent | ScientificName | Family | Author | RankID |
|---|---|---|---|---|---|
| division | Animalia | Mollusca | 30 | ||
| class | Mollusca | Gastropoda | 60 | ||
| order | Gastropoda | Neogastropoda | 100 | ||
| superfamily | Neogastropoda | Buccinoidea | 130 | ||
| family | Buccinoidea | Buccinidae | Buccinidae | 140 | |
| genus | Buccinidae | Buccinum | Buccinidae | 180 | |
| species | Buccinum | Buccinum polaris | Buccinidae | Gray, 1839 | 220 |
Download example CSV View example as a CSV
Tips for preparing your input file
- Parent taxa must already exist in the portal’s thesaurus or be included in the input file. As an alternative to a parent taxon column, as shown above, the input file can contain columns for the core hierarchy that is defined within the taxonunits table (e.g. kingdom, division, class, subclass, order, family, etc).
- Values for rankid are assigned on the backend of your database in the table taxonunits. These value may vary between Symbiota portals. If you do not have access to these values, the Symbiota Support Hub can provide them to you for SSH-hosted portals.
- If a taxon’s acceptance status is not defined (0 = not accepted, 1 = accepted ), all taxa will be assumed to be accepted. Include a separate column in your input file to denote acceptance status, if desired.
- In the absence of a hierarchical definition (e.g. the parent taxa), infraspecific, specific, and genus linkages will be determined from ScientificName and linked to Family. If the Family value does not yet exist in the thesaurus and the hierarchy above Family is not defined in upload field, the taxon will be linked directly to Kingdom.
- You can only batch input taxonomy for one Kingdom at a time (Animalia, Fungi, etc.)
- Make sure not to map more than one source column to the same target, and that source column names are unique from one another.
- The batch loader will import new taxonomy in the order it appears in the file, starting with the first row. It is therefore best to organize the input file such that highest ranks appear first, followed by lower ranking taxa.
- Non-ranked nodes / unranked clades are supported by the bulk loader and do not require rankid values. However, it can be easier to ingest these names using the frontend interface, especially if only a few unranked names are to be added.
- Many fields that appear on Taxon Profile pages can be batch imported using this tool (e.g. vernacular names, language values, etc.). To preview what fields are available for bulk importing, create a small test CSV and open it using the steps 1-4 outlined in the following section.
- Contact the Hub or someone with backend access to add new ranks. Ranks are added on a per-kingdom basis and may require community discussion in order to be inserted into the hierarchy.
Ingest the File Using the Batch Loader
Once your file is prepared, if your user account has the appropriate permissions, you can batch upload taxonomy from a spreadsheet (CSV) or text file by navigating to Sitemap > Taxonomy > Batch Upload a Taxonomic Data File > Taxa Upload.
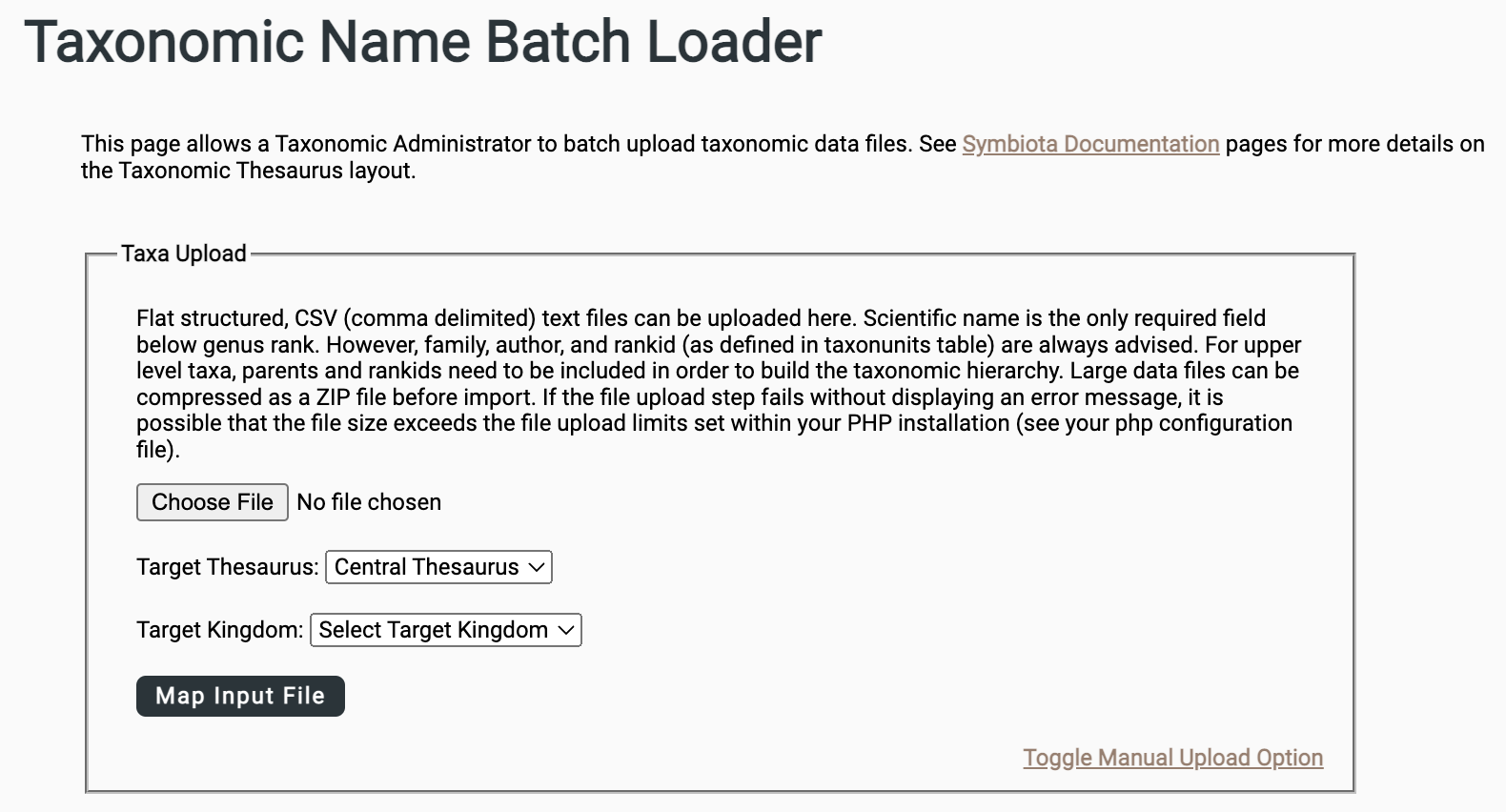
Select your formatted input file.
Select the Target Thesaurus. (Your portal may only have one option.)
Select the Target Kingdom (all names in the input file will be associated with this Kingdom).
Once you select Map Input File, you will be prompted to map the fields in your input file to target fields in the database. Note that the parent taxon = parentstr and scientificName = scinameinput.
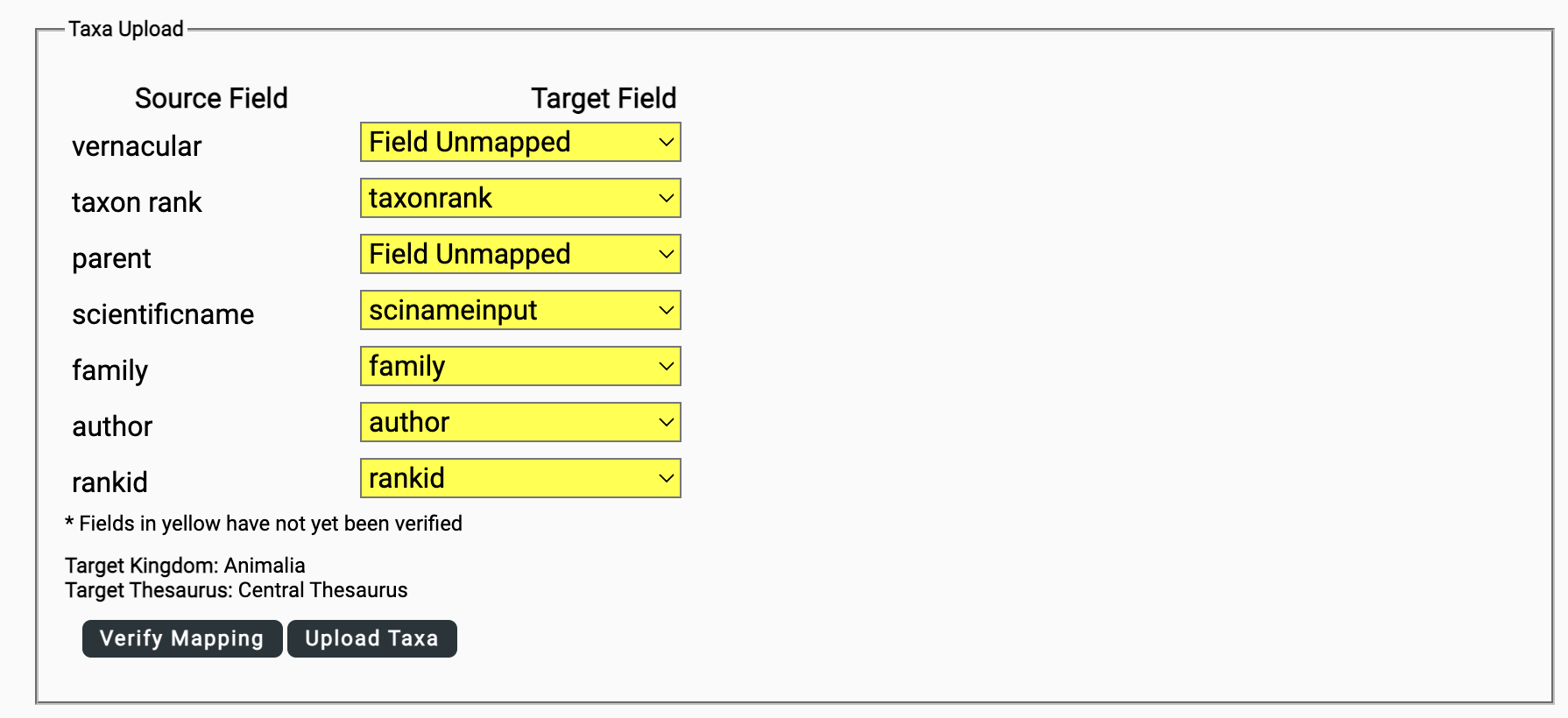
Once the fields are mapped, select Verify Mapping.
Once the file is processed, you will be given the option to review the new names as they were imported into the database’s internal staging tables. The is your last chance to preview changes before they are made permanent in the thesaurus; do so by selecting the link, Download CSV Taxa File. Look for an errorstatus column in the CSV explaining any records that failed to import.
If you are satisfied with the import, select Activate Taxa to transfer the new taxonomy to the portal’s thesaurus. If not, close the window and start over.
Tips for batch importing taxonomy
- If you are new to this process, complete a few test imports to be sure your new taxon records are ingesting correctly. You can review new records in the Taxonomy Explorer (Sitemap > Additional Resources > Taxonomy Explorer).
- For especially complex taxonomic trees, import higher taxonomy first, then import new genera and species-level names.
- When batch importing synonyms, import all accepted names (and, if needed, their higher taxonomy) first. Then import the synonyms and their associated higher taxonomy. A column containing the accepted name for each synonym can be included in your input file.
Cite this page:
Cat Chapman, ed: Lindsay Walker, Katie Pearson. Batch Loading Taxonomic Data. In: Symbiota Support Hub (2025). Symbiota Documentation. https://biokic.github.io/symbiota-docs/portal_manager/taxonomy/batch_load/. Created on 06 Jul 2022, last edited on 22 Jun 2023.