Importing Determinations
This page provides instructions for bulk uploading specimen determinations/annotations and associating them with their respective specimen records. For instructions for adding individual determinations or the batch annotation tool, see this page. This tool can only be accessed by a superadministrator.
If you are new to using this tool, try importing only a few new determinations to familiarize yourself with how the tool behaves before ingesting a large batch. For instance, this tool cannot be used to overwrite existing determinations; it can only be used to add new determinations to one or more occurrence records.
 |
|---|
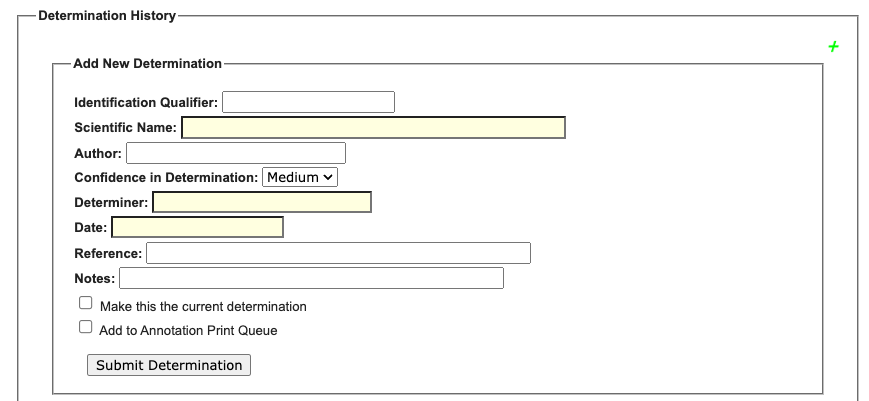
| Data associated with specimen identifications (aka taxonomic determinations or annotations) can be added to specimen records in bulk using the Extended Data Importer tool. |
Initiating the Upload
- Navigate to your Administration Control Panel (My Profile > Occurrence Management > name of your collection).
- Within the Administration Control Panel, navigate to Import/Update Specimen Records > Extended Data Import.
- Select the file containing your determinations to be imported. See below for tips regarding the contents and formatting of this file.
- Choose Import Type = “Determinations”.
- Select the desired Upload Type from the dropdown menu.
- If your upload was successful, the message “Done process input file” will appear in the Action Panel.
Example Data Import File
Files containing determinations to be bulk imported must be in CSV format and include the following: 1) an identifier for the associated specimen occurrence (catalogNumber, otherCatalogNumber, or occurrenceID), 2) dateIdentified, 3) identifiedBy, and 4) sciname.
For example:
| catalogNumber | identifiedBy | dateIdentified | sciname |
|---|---|---|---|
| OBI000001 | Don Cole | 1950 | Rosa woodsii var. gratissima |
| OBI000001 | unknown | s.d. | Rosa |
| OBI000002 | Edward Gilbert | 2023-10-01 | Rosa woodsii |
Data Import Fields
Refer to the Symbiota Data Fields and Data Import Fields guides for more information about some of the fields listed below.
Not all fields available through the Extended Data Importer for bulk determination ingestion are visible through the Occurrence Editor interface. However, once imported, these values can be exported, for example, through a data backup.
| Target Field (🔸 = required) | Data Type (Length) | Notes |
|---|---|---|
| subject identifier 🔸 | catalogNumber, otherCatalogNumber, or occurrenceID | |
| dateIdentified 🔸 | Text (45) | The date the identification was made. If the date is unknown, use “s.d.” in this field. |
| family (Override) | Text (255) | The family to which the taxon belongs. If the scientific name provided is matched to one in the taxonomic thesaurus, this will be auto-filled according to the thesaurus entry. |
| higherClassification (Override) | Text (150) | A list (concatenated and separated) of taxa names terminating at the rank immediately superior to the referenced dwc:Taxon. If the scientific name provided is matched to one in the taxonomic thesaurus, this will be auto-filled according to the thesaurus entry for the current identification. |
| identificationQualifier | Text (255) | The determiner’s expression of uncertainty in their identification. This will be listed on the label along with the scientific name. (e.g., cf., aff.) |
| identificationReferences | Text (2000) | The reference source used to make the identification. |
| identificationRemarks | Text (2000) | Any additional notes regarding the identification of the specimen. |
| identificationUncertain | Integer (2) | Use the value “1” to indicate an uncertain identification or “0” for certain identifications. |
| identificationVerificationStatus | Text (45) | A categorical indicator of the extent to which the taxonomic identification has been verified to be correct. |
| identifiedBy🔸 | Text (255) | The name of the person who identified the specimen. Also called a determiner. If the determiner is unknown, use “unknown” in this field. |
| isCurrent | Integer (2) | Use the value “Y” to flag a determination as the most current. However, doing so will not override existing determinations that have already been flagged as the most current. |
| printQueue | Integer (2) | Use the value “1” to add a determination to the annotation print queue. |
| scientificNameAuthorship | Text (255) | The name of the person who first named the taxon. If the scientific name provided is matched to one in the taxonomic thesaurus, this will be auto-filled according to the thesaurus entry. |
| sciname 🔸 | Taxonomic identification without authorship string. See Data Import Fields. | |
| securityStatus | Integer (11) | Use the value “1” to redact images and locality data for the associated occurrence record; use “0” to keep all details visible. See Symbiota Data Fields. |
| securityStatusReason | Text (100) | Explanation for data redaction. See Symbiota Data Fields. |
| sortSequence | Integer (10) | Specifies the sequential sort order of multiple determinations on the determination history tab and in the public display. Low numbers will sort higher than high numbers. |
| sourceIdentifier | Text (45) | An identifier for the Identification (the body of information associated with the assignment of a scientific name). May be a global unique identifier or an identifier specific to the data set. |
| taxonConceptID | Text (45) | An identifier for the taxonomic concept to which the determination refers. |
| taxonRemarks | Text (2000) | Any additional notes regarding the taxonomic name to which the specimen was identified. |
| verbatimIdentification | Text (250) | A string representing the taxonomic identification as it appeared in the original record. |
Cite this page:
Lindsay Walker, ed: Katie Pearson. Importing Determinations. In: Symbiota Support Hub (2025). Symbiota Documentation. https://biokic.github.io/symbiota-docs/portal_manager/determinations/. Created on 31 Oct 2023, last edited on 06 Jun 2024.